Zlab 发表文章
Explore ZLab's recent research articles and collaborations.
2025
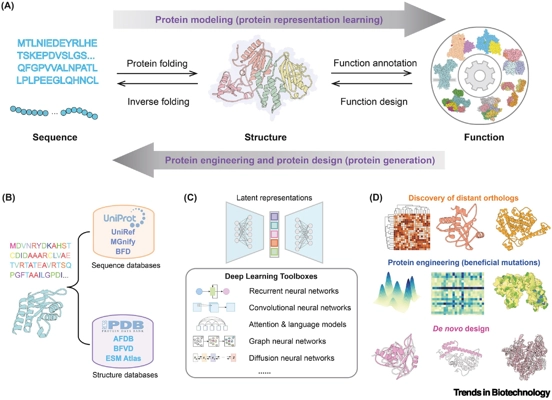
AI sheds new light on genome editing
Trends in Biotechnology
2024
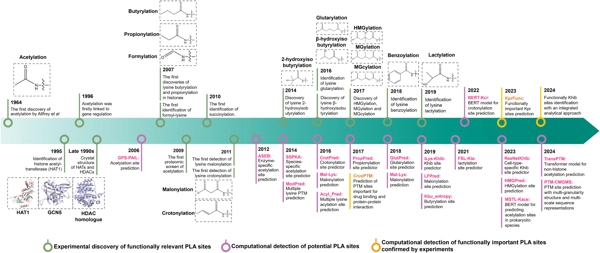
Current computational tools for protein lysine acylation site prediction
Briefings in Bioinformatics
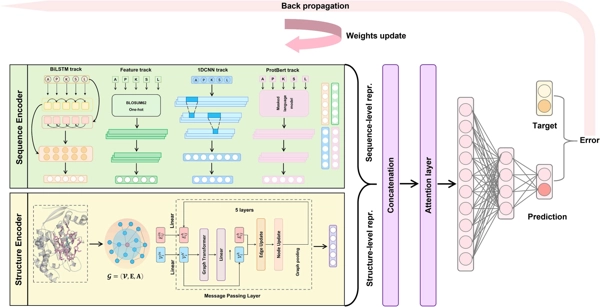
SLAM: Structure-aware lysine β-hydroxybutyrylation prediction with protein language model
International Journal of Biological Macromolecules
2023
2022
2021
2020
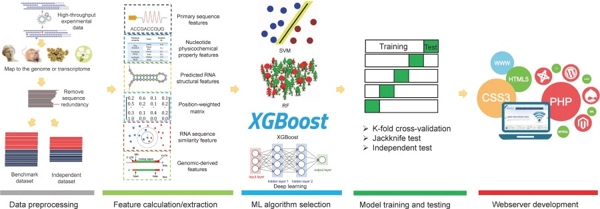
Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences
Briefings in Bioinformatics, Volume 21, Issue 5, Pages 1676–1696
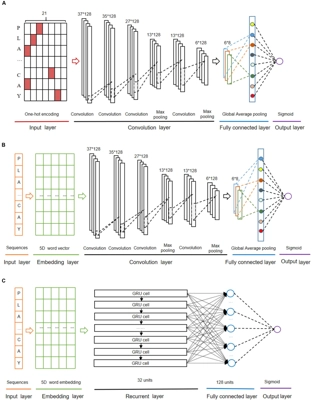
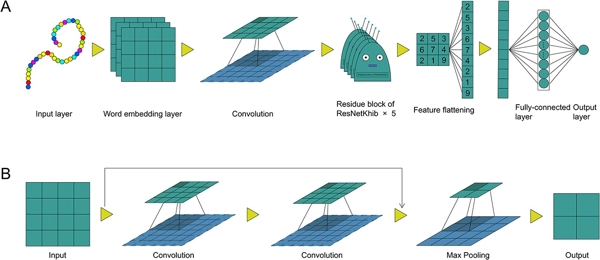
DeepKhib: A Deep-Learning Framework for Lysine 2-Hydroxyisobutyrylation Sites Prediction
Front. Cell Dev. Biol.
2019
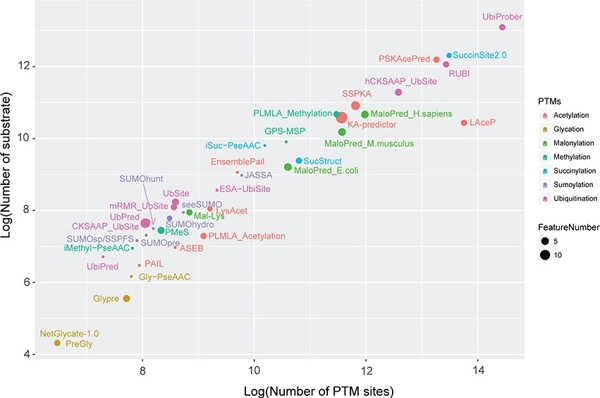
Large-scale comparative assessment of computational predictors for lysine post-translational modification sites
Briefings in Bioinformatics, Volume 20, Issue 6, Pages 2267–2290
2018
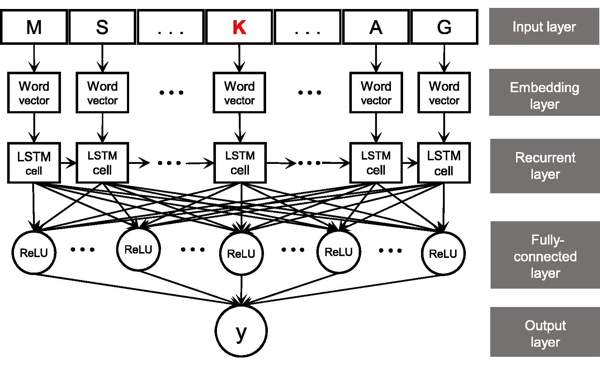
Integration of A Deep Learning Classifier with A Random Forest Approach for Predicting Malonylation Sites
Genomics, Proteomics & Bioinformatics, Volume 16, Issue 6, Pages 451–459

iFeature: a Python package and web server for features extraction and selection from protein and peptide sequences
Bioinformatics, Volume 34, Issue 14, Pages 2499–2502