Introduction to lysine acylation!
As a main subtype of post-translational modification (PTM), protein lysine acylations (PLAs) play crucial roles in regulating diverse functions of proteins. With recent advancements in proteomics technology, the identification of PTM is becoming a data-rich field. A large amount of experimentally verified data is urgently required to be translated into valuable biological insights.
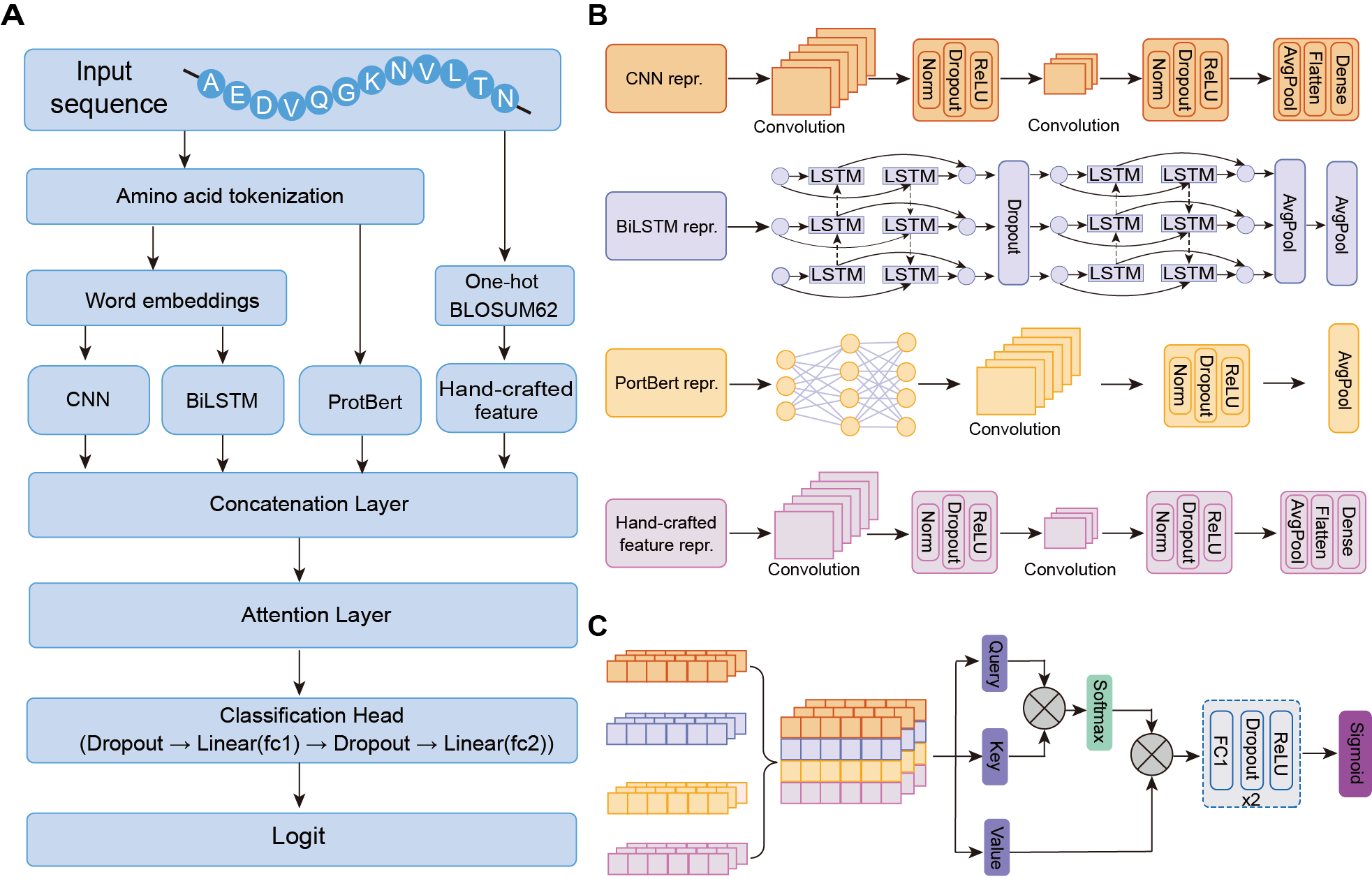
Hybrid deep learning model architecture
The developed hybrid deep learning framework includes 1) a multi-track encoder module to concurrently embed the sequence features into a latent representation; 2) a decoder layer consisting of an attention layer and a multi-layer perceptron followed with a sigmoid function for downstream classification. The sequence encoder is designed as hybrid deep learning neural networks to learn dependencies between residues with two-track feature encoders and two-track adaptive encoders. Adaptive encoders enable learn-from-data for RicePLA by using learnable word embeddings, and feature encoders provide expert-level information and evolutionary constraints extracted from protein language model.
If you use RicePLA in your work or publication, please kindly cite the following paper:
M. Zhang et al., RicePLA: An integrated deep learning framework for accurate prediction of multitype rice lysine acylation sites
Qin, Z., Ren, H., Zhao, P., Wang, K., Liu, H., Miao, C., Du, Y., Li, J., Wu, L., & Chen, Z. (2024). Current computational tools for protein lysine acylation site prediction. Briefings in bioinformatics, 25(6), bbae469.